

On Wednesday 17th February 2021 the ESCAPE consortium held the “Enhancing science through sharing software - benefits & use cases” webinar with the aim to share the ESCAPE Open source Scientific Software and Service Repository latest development status, best practices and use cases on how to enhance scientific discoveries by sharing software and fostering software co-development. The webinar gathered 62 registrants, 88% of which coming from an Academia/research background.
ESCAPE Open source Scientific Software and Service Repository prototype
After giving a general overview of the ESCAPE project and its relation with the European Open Science Cloud, Kay Graf, the ESCAPE OSSR WP3 leader, explained how ESCAPE is working to make software and derivatives of software openly available based on the FAIR principles to foster and enable open science, software interoperability and re-use, federation of available software resources and cross fertilisation between the European Strategy Forum on Research Infrastructures (ESFRIs) and beyond.
Figure 1 – ESCAPE OSSR prototype
Kay then shared how to improve the ESCAPE OSSR onboarding process. There is constant coordination between the EOSC core, established community platforms and solutions and the ESCE OSSR, consisting of a landing page, a development platform (GitLab instance) and a repository (Zenodo).
Figure 2 – ESCAPE OSSR development platform and repository
During the presentation, the “Crab bundle” was introduced as an example of open science. The crab bundle use case is an example of an open science project using already available open data to perform a re-analysis, which has been made available on the Zenodo public repository. This use case has been made possible thanks to an infrastructure and services that host FAIR contributions:
- Findable: Data is described with rich metadata and assigned a unique and persistent identifier.
- Accessible: Metadata identifiers follow standard (open, free, universal) communication protocols – accessible even when data is no longer available.
- Interoperable: Metadata uses a formal, accessible, shared, and broadly applicable language for knowledge representation (metadata schema or standard).
- Reusable: Data and collections have clear usage licenses and provide accurate information on their source.
Common software development use cases: benefits for the users
There are already some use case projects developed under ESCAPE using the Open Source Scientific and Software Repository, such as:
- Agnpy: Agnpy is a python tool that aims to provide a library to compute multi-wavelength photon spectra produced by leptonic radiative processes in jetted Active Galactic Nuclei. This library allows other researchers to reproduce the models and the spectra that have been produced in the past and build new and more complex models on top of them.
- IndexedConv: IndexedConv performs deep learning convolution and pooling operations on non-Euclidian grids of data as an alternative to masking or resampling the data. It is a generic solution that allows researchers to apply convolution to unconventional images, such as those taken with a camera that does not have the traditional shape and has hexagonal pixels.
- ConCORDIA: ConCORDIA is a set of containers for CORSIKA simulations on DIRAC. It will allow researchers to find the ready to use container for their CORSIKA simulations. Some of the use cases for this project are KM3NeT, CTA, etc..
It will be released soon in the ESCAPE OSSR Zenodo repository. - gLike: Phyton code general-purpose ROOT-based code framework for the numerical maximisation of joint likelihood functions. It can be used to estimate energy scale of quantum gravity, dark matter annihilation cross-section, etc..
- Machine learning and pipeline developments for CTA: Deploying a framework for CTA to apply deep learning to find the gamma event type, its energy and duration from a set of raw data. This use case allows other researchers to re-run these experiments.
The fully documented source code of these onboarded ESCAPE OSSR use cases can be found in the OSSR Zenodo repository with an open license that will allow other researchers to use and modify them for their own purposes.
SKA Data challenges towards full reproducibility
The Square Kilometre Array has designed data challenges to represent the workflow of users interacting with their data products to help promote software development workflow best practices regarding:
- Coding standards: Style guide, documentation, comments, functions, classes, modular and version control.
- Publishing their code, data and workflow alongside the results.
- Ensuring their workflow is reproducible.
Promoting full replication of code and data is the gold standard in any scientific data analysis, as a way to simplify scientific discoveries.
Without this replicability, scientists will have to go through extensive effort to replicate certain results from a specific dataset, which is a clear obstacle for open science scientific innovation.
On the other hand, promoting replicability best practices, such as sharing containers with versioned data, allows other researchers to quickly replicate any research results and potentially spark new scientific breakthroughs.
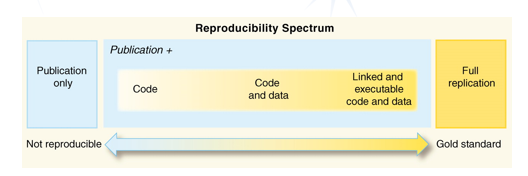
Figure 3 - Peng 2011 (https://doi.org/10.1126/science.1213847)
(and Rachael Ainsworth, https://github.com/rainsworth/osip2019-containerisation-workshop)
Given the growing amount of data produced by recent researchers, such as SKA’s researchers, it is extremely important to apply replicability best practices to any scientific analysis made with such data, for the benefit of all researchers and society at large.
Innovative machine learning workflow for real time multi-messenger analysis
Astro and particle physics experiments are producing an incredible and increasing amount of data, which is making it difficult for researchers to easily access data archives, label objects in an astrophysical catalog and classify noise or signal sources without human intervention. Before the amount of data becomes too big to be managed by current algorithms, it is critical to develop new workflows for data analysis. The ESCAPE partners, namely KM3NeT-ORCA/ARCA, the Square Kilometre Array and the Cherenkov Telescope Array are already implementing such innovative workflows using machine learning to help them navigate the growing amount of data produced.
ESCAPE Open source Scientific Software and Service Repository: what is next!
The webinar ended with Kay Graf sharing a sneak peek into the future of the ESCAPE OSSR service. The ESCAPE team is expected to finalise the OSSR service prototype implementation to foster software co-development, re-use and innovation and create its full integration into the EOSC Portal by August 2022.
Follow us on Twitter
Subscribe to our newsletter
See the webinar video below
Views
19,301

